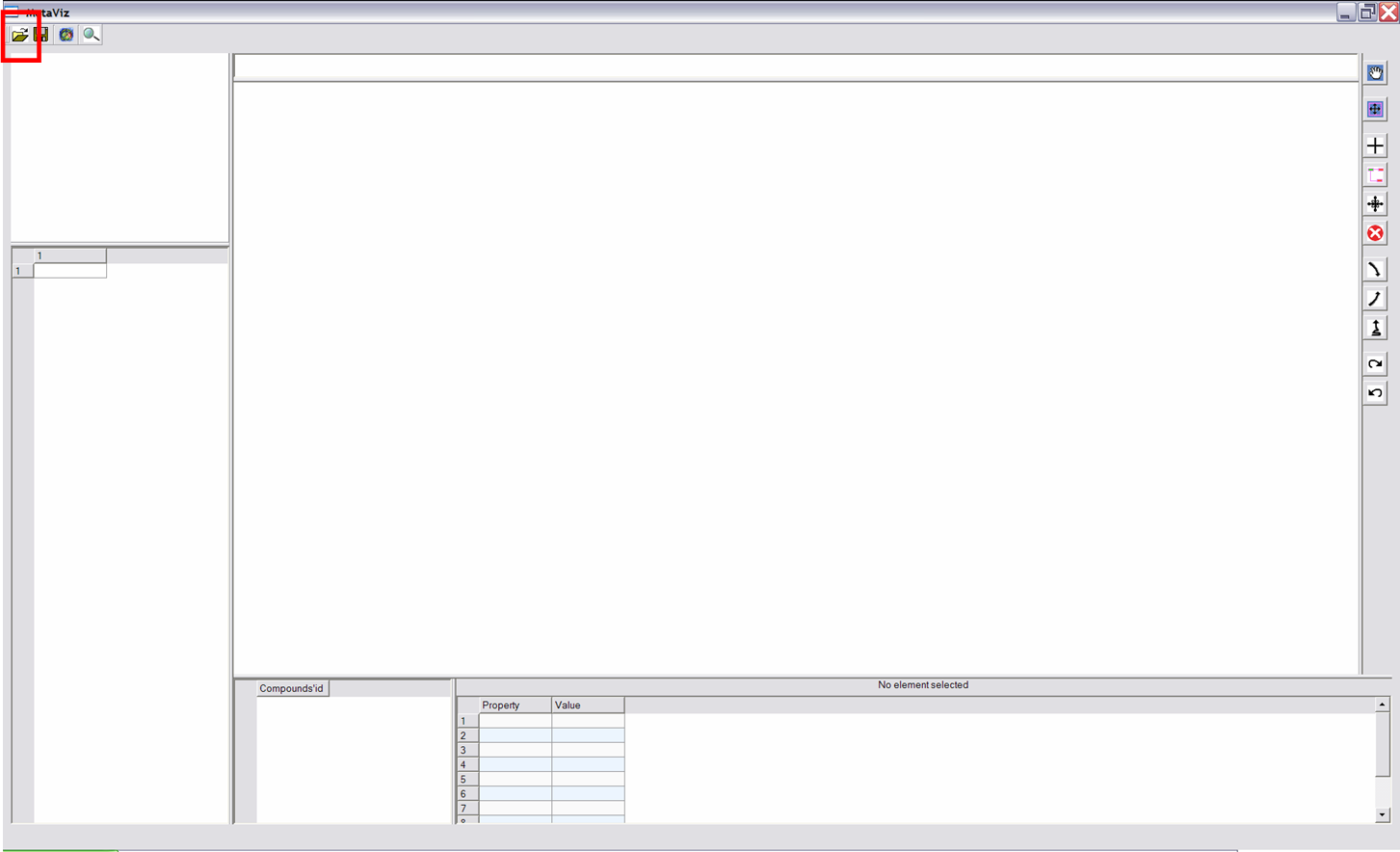
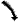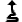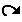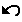Interaction and Navigation Buttons
Each time, one clicks on an interaction button, effects of left/right button and wheel of mouse change.
| Navigation |
 |
After clicking on this button, the mouse allows to navigate in the drawing : left button to "move" and wheel to zoom in or out. |
 |
This button allows to go down in the hierarchy. Click on a metanode to focus on nodes it contain. |
 |
This button allows to go up in hierarchy. Click on it to go up of one level. |
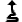 |
This button allows to go up in hierarchy. Click on it to go to the upper level of the hierarchy. |
| Selection |
 |
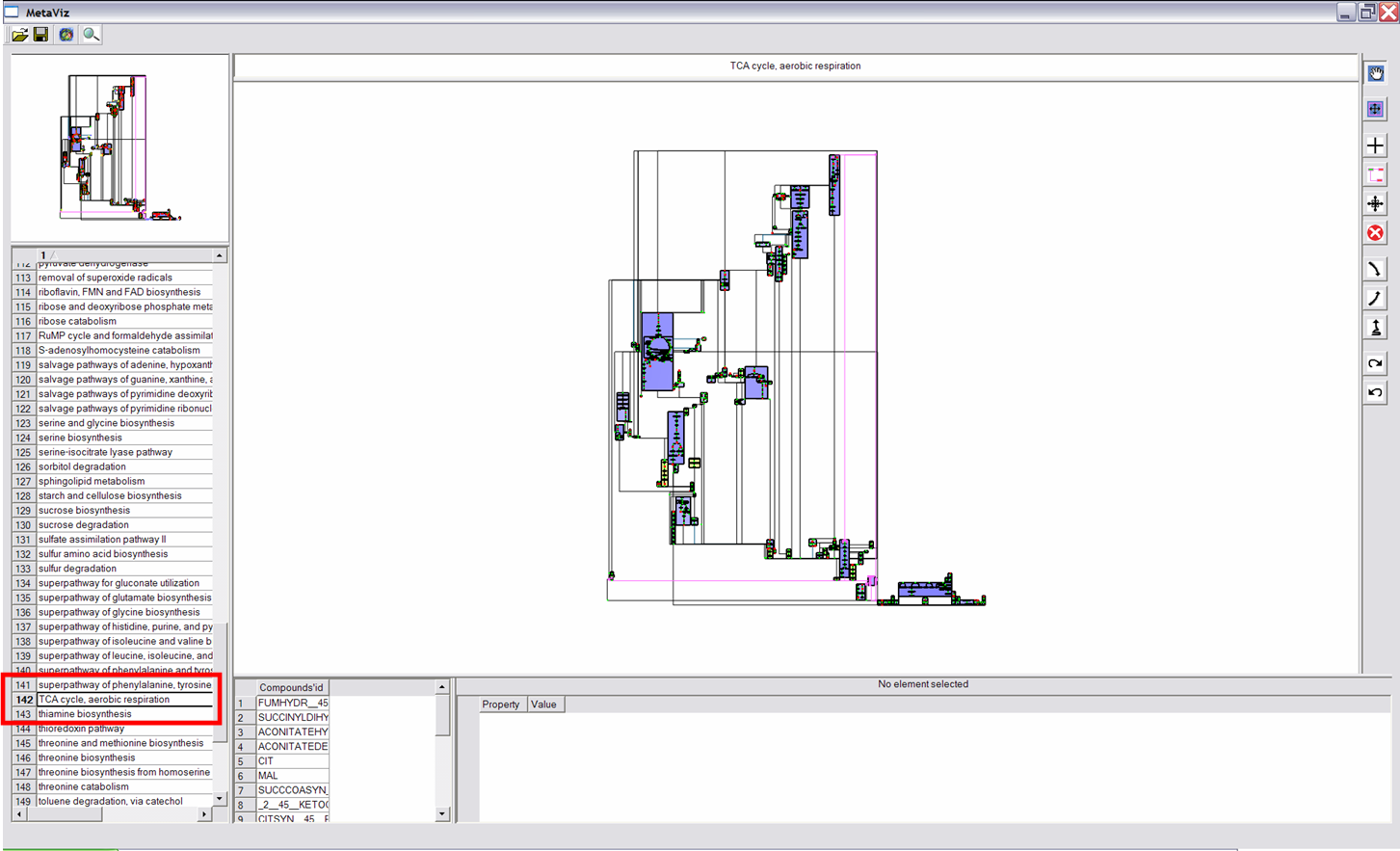
This button allows to select a node or a metanode, when cliking on a node (using left button), it is highlightened in pink. Properties of the node are described on the top (if it is a metabolic pathway) and on the bottom. |
 |
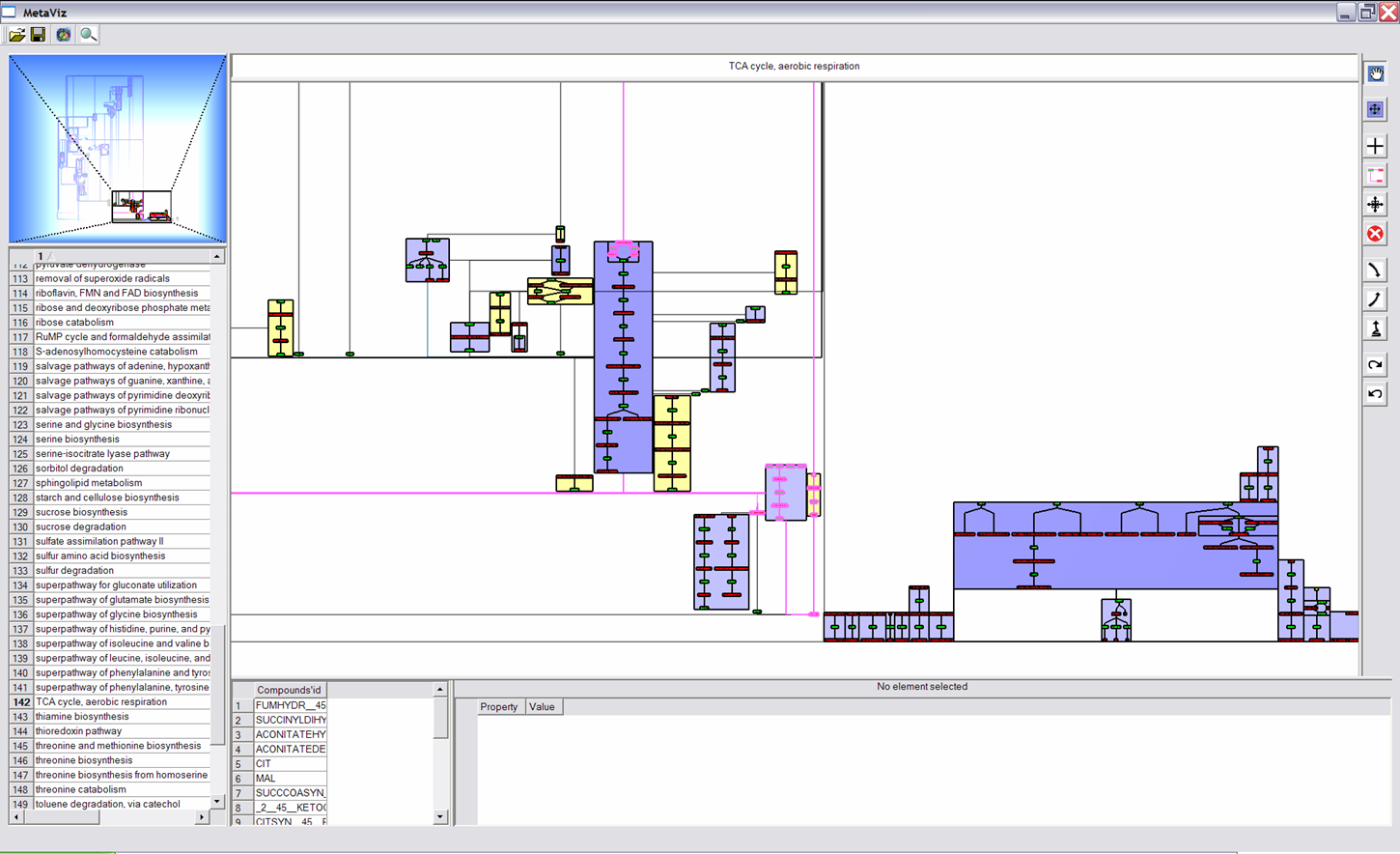
This button allows to visualize the neighborhood of a node. When clicking on a node (using left button), it is highlightened and so are its neighbors. If the clicked node is a metanode, all nodes (and all their neighbors) in it are highlightened. |
 |
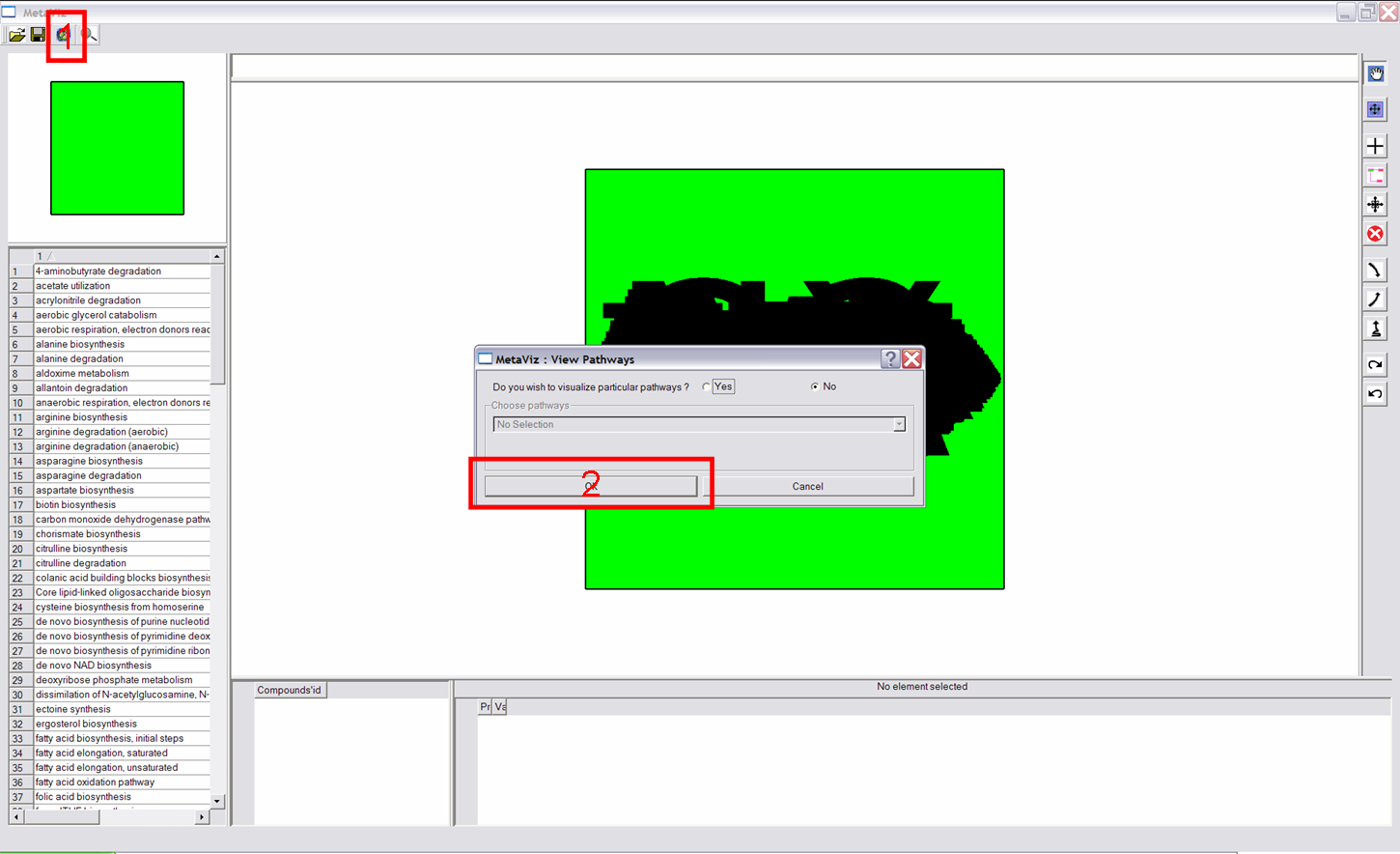
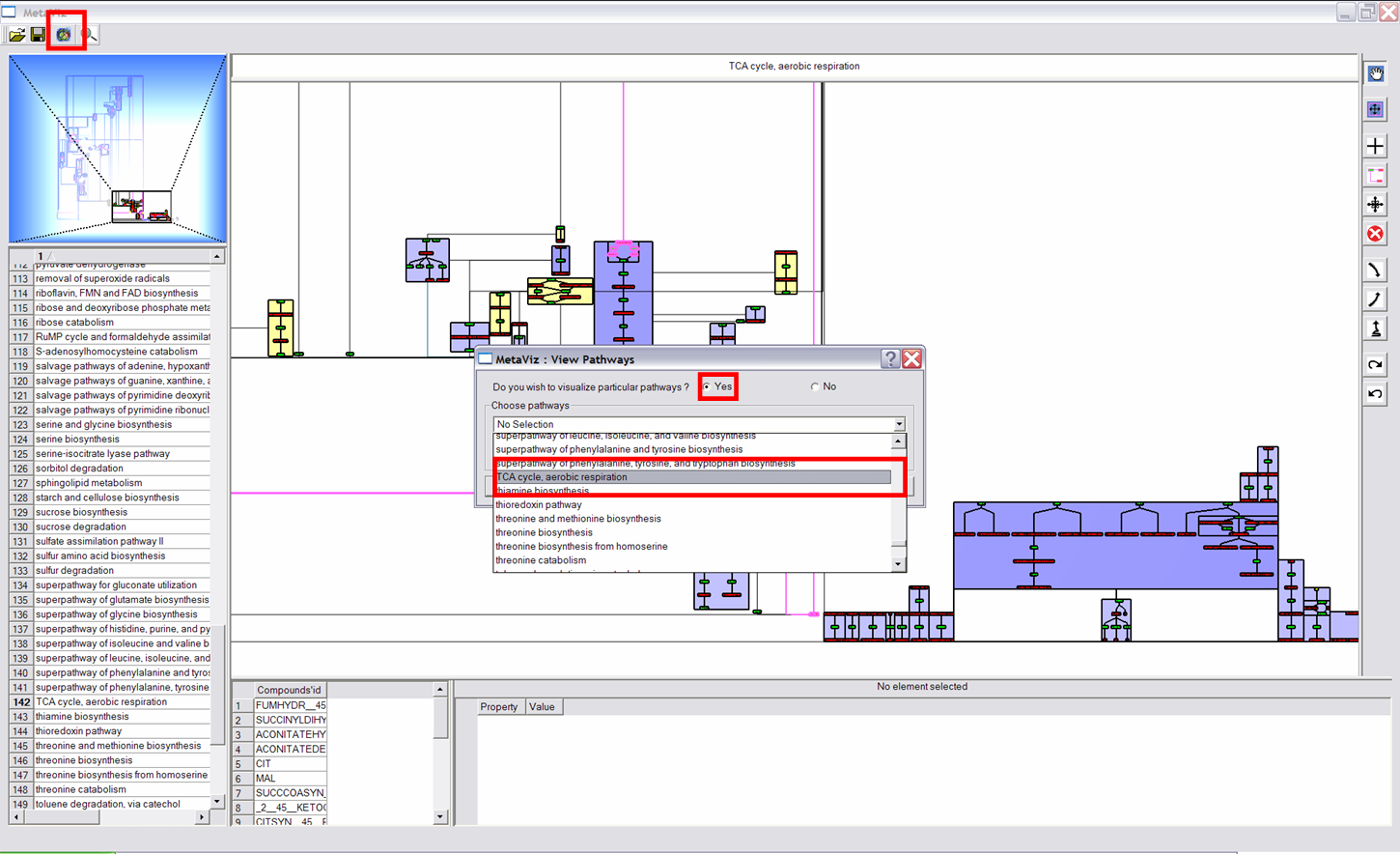
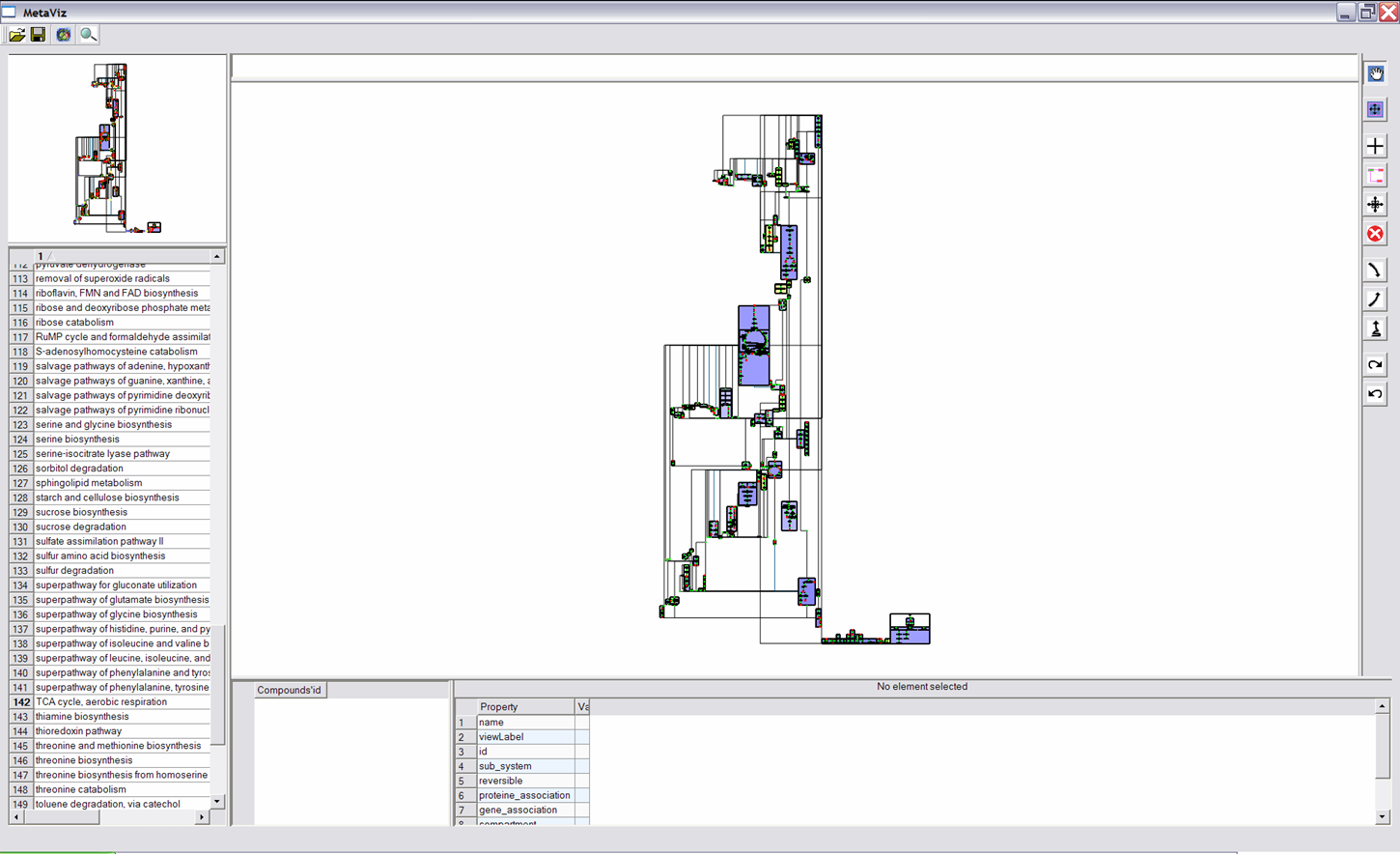
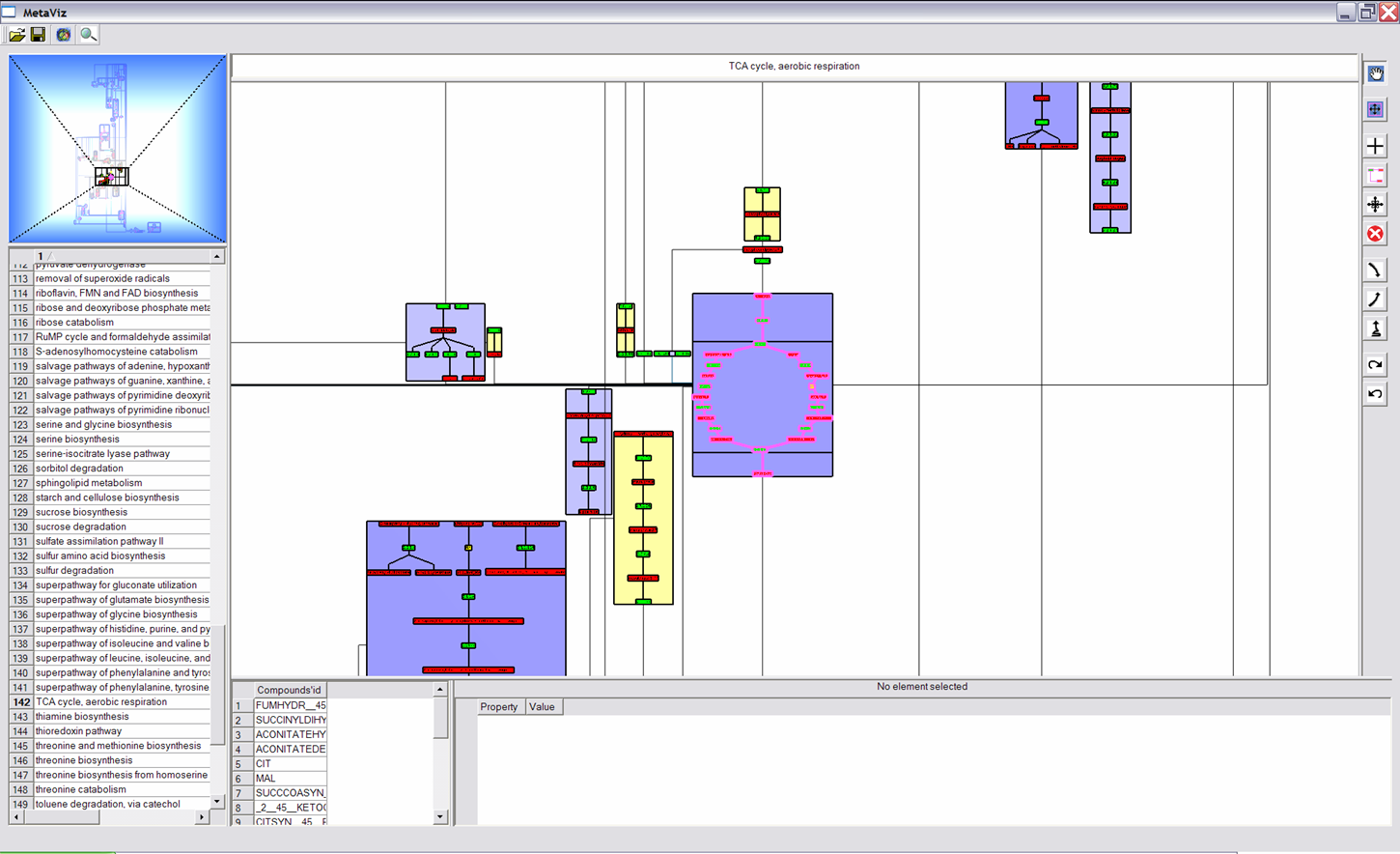
This functionality allows to visualize each metabolic pathway a node (compound) belongs to. First click on a node, then scroll the wheel to see pathways it belongs to (name of these pathways are written on the top and composition of these pathways are shown on the bottom). |
 |
This button allows to unselect all nodes. |
| Interaction |
 |
This functionality allows to change the layout of nodes. To move a node, you first have to "select" it using the cross button described below, then left-click on the selected node, drag it and release at its new position . |
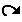 |
After selecting a metanode (using the selection interactor), click on this button to open a metanode. |
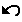 |
This button allows to "re-group" the lastest opened metanodes. |
 MetaViz Tutorial
MetaViz Tutorial