 CoPHI
CoPHI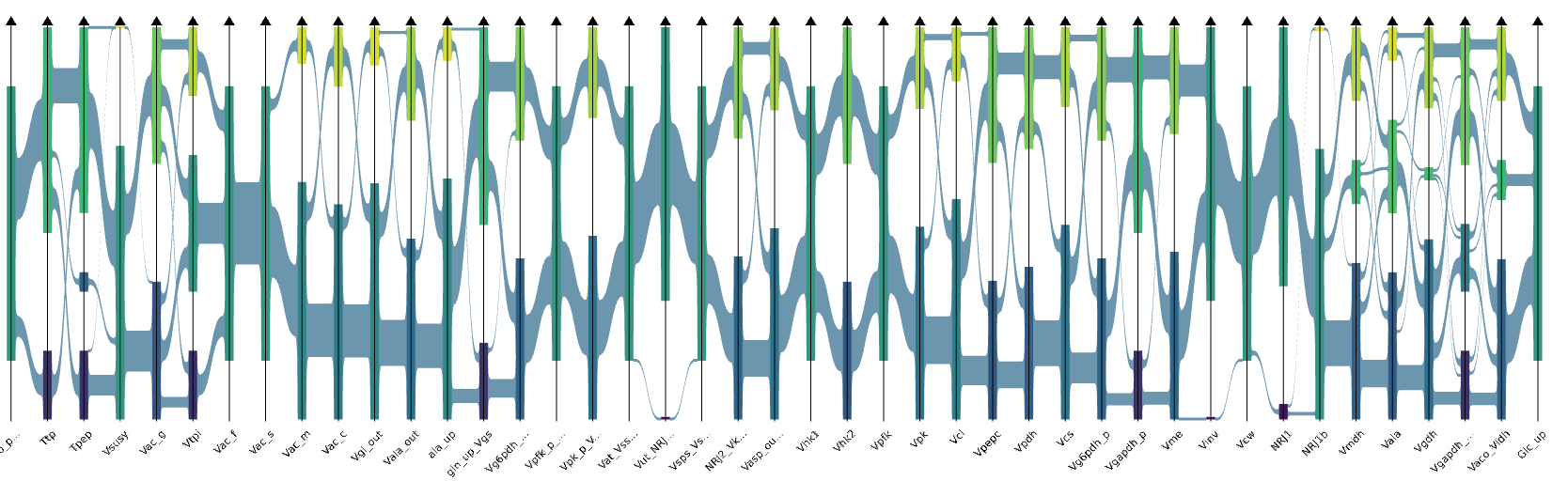
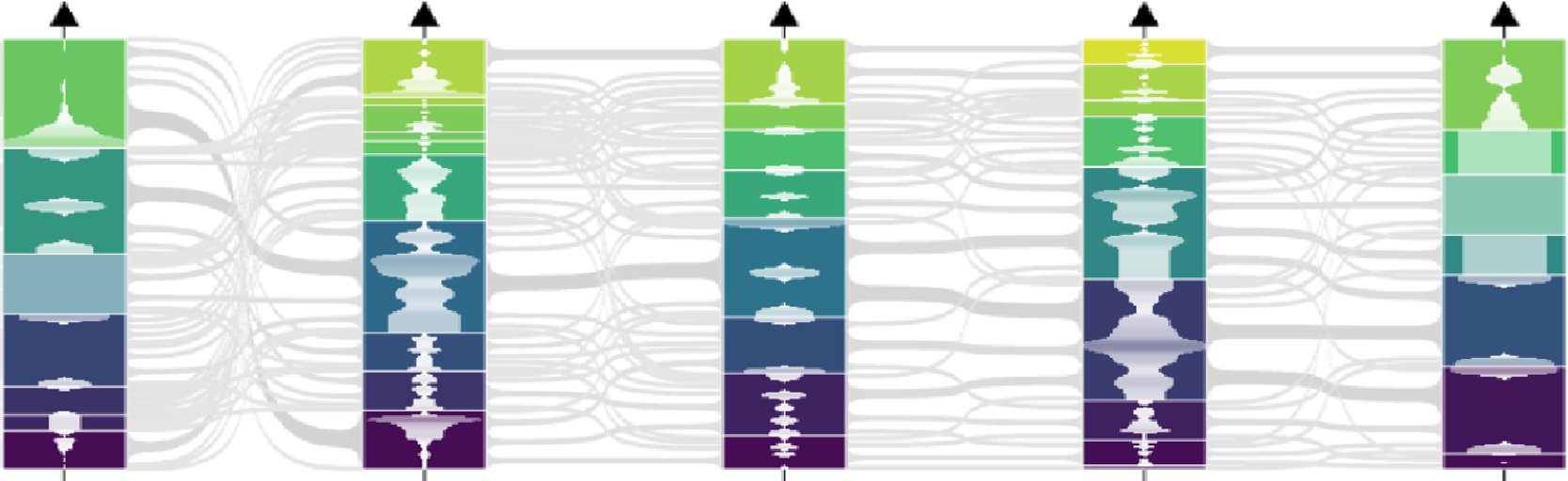
CoPHI is an interaction-ready parallel coordinates visualization system. Originally designed from works on visualization and interactive exploration of Big Data using data abstraction.
Still in development, this system have been designed and tested with Mozilla Firefox; thus we cannot ensure the proper functioning with other web browser.
Finding reaction patterns through metabolic pathways allows to identify constraints and interaction between enzymatic reactions.
Elementary flux modes computing is a way to know
feasible pathways through a metabolic network. The set size
of these feasible pathways requires tools to display the
results. CoPHI is a user-friendly web application to
display the set of Elementary Flux modes by using
parallel coordinates graph.
To test with a concrete example coming from analysis of
plant metabolism follew this link .
To cite CoPHI, please refer to the original article :
Sansen, J.; Richer, G.; Jourde, T.; Lalanne, F.; Auber, D.; Bourqui, R. "Visual Exploration of Large Multidimensional Data Using Parallel Coordinates on Big Data Infrastructure". Informatics 2017, 4, 21.
BioNetVisA 2016 : Metabolic Networks: Visual Analysis of Elementary Flux Modes.
GT Bioss - Montpellier 2017 : How to display patterns inside elementary flux modes.