What a pleasure it was to attend (as a guest) Maxime Mahout’s thesis, entitled ‘Logic programming for the analysis of metabolic flows and applications to biology’, supervised by Sabine Pérès. It’s a thesis that I’ve been involved in since Maxime’s Master 1 course (yes!), through 5 years of discussions, covid visios, crazy ideas, moments of doubt and discoveries (for my part) of cellular mechanisms and frightening diseases. The idea is to counterbalance the incredible combinatorial complexity of the mechanisms of cells (and even consortia of cells) with the power of logical reasoning and constraints. Thanks to the specialisation of an ASP engine, maxime has been able to propose avenues of treatment for diseases that are still dramatic, by demonstrating unfailing perseverance.
We have therefore been able to take advantage of the tremendous modelling work carried out by biologists, making the mechanisms at work in cells accessible through logic (or in the form of graphs). This makes it possible to reason about the mechanisms at work in cells, which are seen as metabolite factories, producing proteins from clearly identified chemical rules. Once this modelling has been done, many problems can be formulated in logic (which gene should be deactivated to prevent the cell from producing a given enzyme, while retaining its good properties?), opening the way to new analytical possibilities.
I can only advise you to read the thesis, or to refer to the article we were able to publish in Nature Processing Journal (Systems Biology and Applications), by following this link.

Maxime’s thesis work has led to the development of therapies for extremely serious infections that cause thousands of deaths and amputations. He has identified what is special about two bacteria (which can be treated individually) working together to survive. When P. aeruginosa and S. aureus colonise a wound, it is virtually impossible to treat the infection, as the consortium becomes extremely resistant to all antibiotic treatments. In his work, Maxime has even managed to identify the metabolic pathways that need to be inhibited to potentially block the entire consortium.
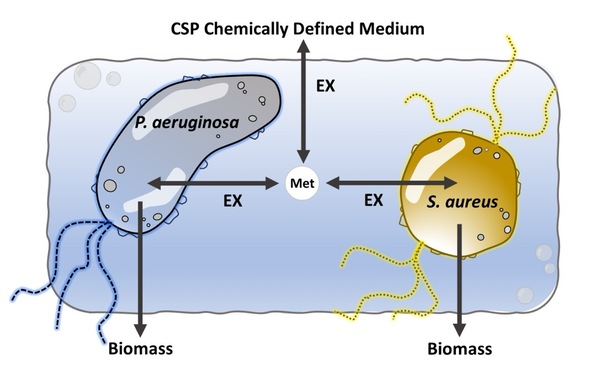 An illustration of the bacteria P. aeruginosa and S. aureus, which, when they work together, become absolutely formidable to combat.
An illustration of the bacteria P. aeruginosa and S. aureus, which, when they work together, become absolutely formidable to combat.
For me, this was an opportunity to appreciate the importance and difficulty of modelling in biology, the extreme combinatorial nature of metabolic networks and the strength of logical reasoning, which is the only way to grasp this complexity and see clearly, in a framework far removed from the resolution of random or toy instances, which are often the main objects in front of the researcher at a very early stage.
Well done again Maxime!